Previous Projects
This page details my previous research projects that have significantly shaped my skills and interests. Each project highlights my contributions to hypothesis development, data analysis, and publication, demonstrating a strong foundation in both molecular biology and computational research.
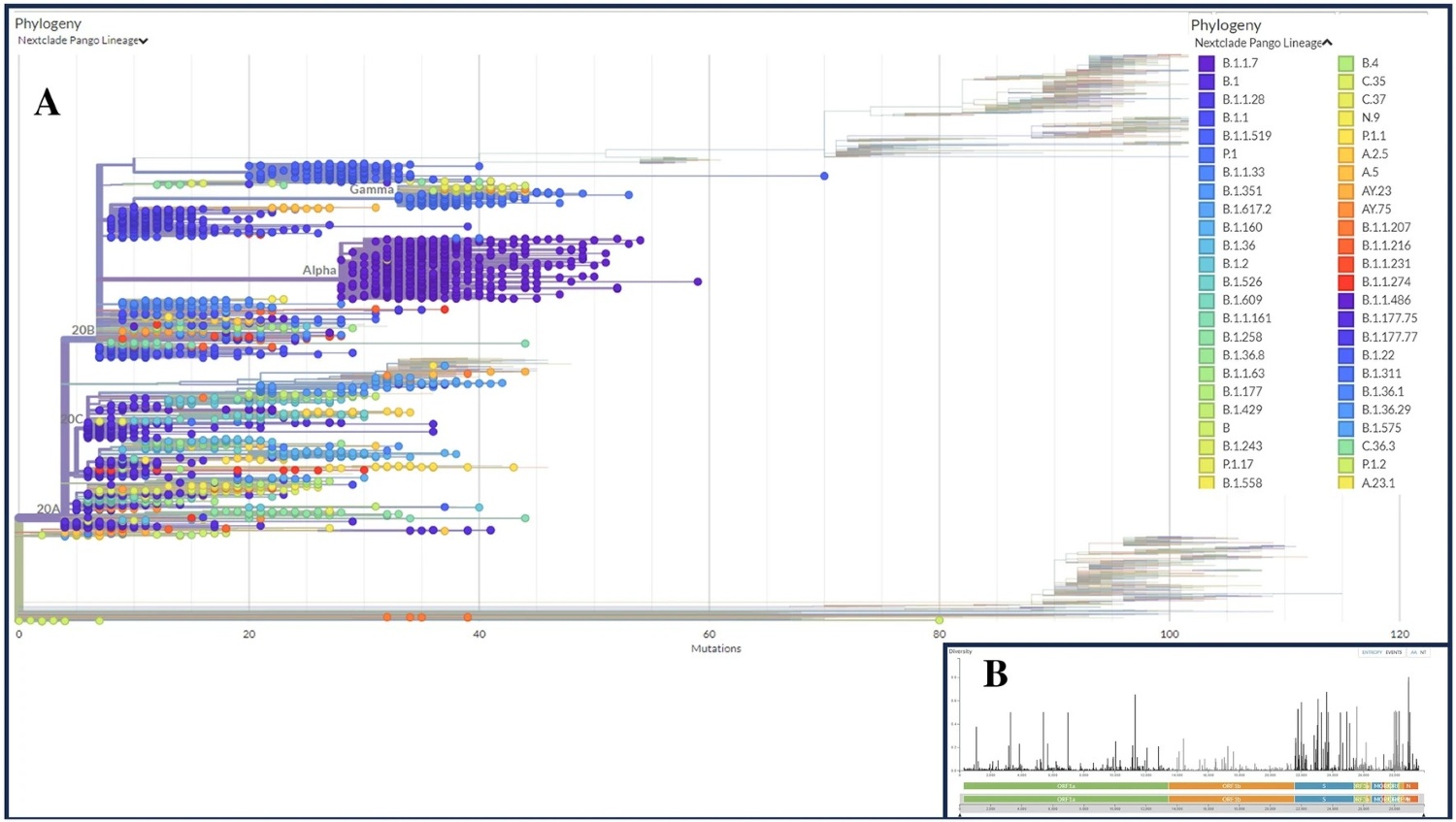
1. SARS-CoV-2 Mutational and Immunoinformatic Profiling
Role: Research Assistant
Institute: Department of Biochemistry and Molecular Biology, SUST
Duration: April 2021 – January 2023
This project focused on mutation profiling and immune landscape mapping of SARS-CoV-2 variants, including Delta and Omicron. Using a dataset of over 200,000 GISAID sequences, I applied tools like NextClade, PredictSNP, and DynaMut to identify deleterious mutations in the spike protein, including those associated with immune escape and protein destabilization.
Machine learning models were trained to classify high-risk mutations, integrating immunoinformatics tools (IEDB, NetCTLpan) to assess T-cell and B-cell epitope variability. This resulted in several impactful publications:
- Scientific Reports (2023): Analysis of shared mutations across deceased patients globally
- Frontiers in Pharmacology (2023): Variant-specific mutation mapping for vaccine strategy
- Advances in Biomarker Sciences and Technology (2023): Multi-omics biomarker analysis in cancer–diabetes overlap
- Frontiers in Immunology (2022): Natural immune enhancers during COVID-19
I performed data preprocessing, statistical modeling in R and SPSS, and manuscript figures using Adobe Illustrator.
2. Wastewater-Based SARS-CoV-2 Surveillance & Environmental Virology
.jpg)
Role: Research Assistant
Institute: Department of Civil & Environmental Engineering, SUST and COVID-19 Testing Lab at NSTU
Duration: November 2020 – June 2021
This interdisciplinary project applied environmental sampling and molecular diagnostics to detect SARS-CoV-2 RNA in wastewater from communities with limited sanitation infrastructure. As part of the project:
- I collected and processed wastewater samples.
- Extracted RNA and performed qRT-PCR.
- Developed a monitoring framework using R for spatial and temporal trends.
This work led to the following publications:
- Environmental Pollution (2022): Surveillance protocol for low-resource settings
- Current Opinion in Environmental Science & Health (2023): Integration of wastewater data with clinical diagnostics
- medRxiv (2021): Pilot studies on decentralized wastewater monitoring
These efforts contributed directly to early warning models for COVID-19 outbreaks and showcased my ability to work across microbiology, public health, and data science domains.
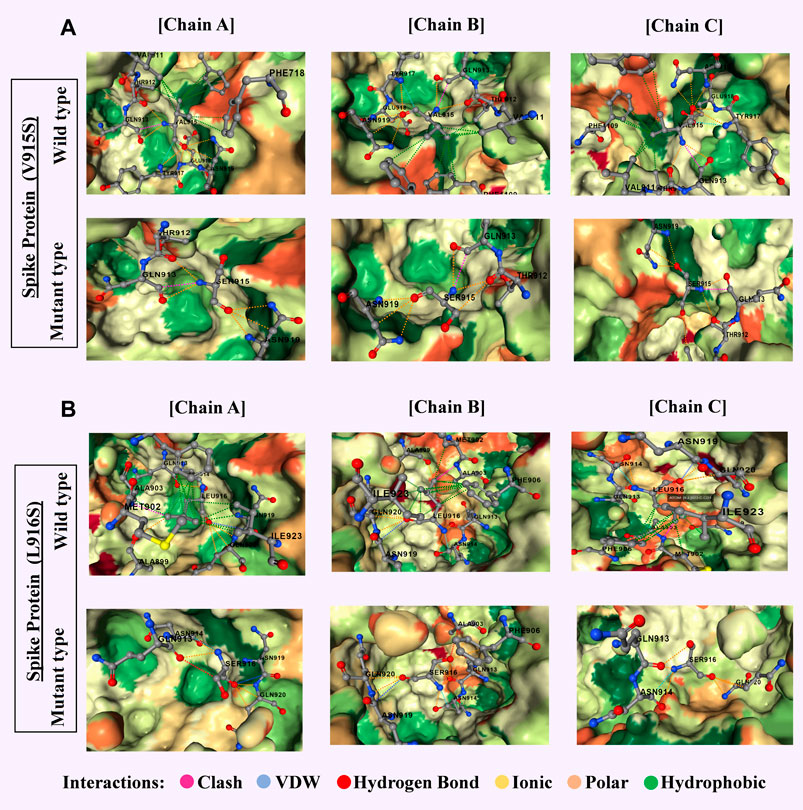
3. Molecular Docking and MD Simulation for SARS-CoV-2 RBD Inhibitors
Role: Research Assistant (Remote)
Institute: Red-Green Research Center
Duration: February 2020 – December 2021
Using structure-based drug discovery techniques, this project aimed to design small molecules and RNA aptamers that could bind to the SARS-CoV-2 receptor-binding domain (RBD) with high affinity. I used YASARA, GROMACS, and Desmond for MD simulations and docking validation.
- Curated ligand libraries.
- Modeled RBD-ligand interactions.
- Ran Linux-based simulation pipelines for conformational stability.
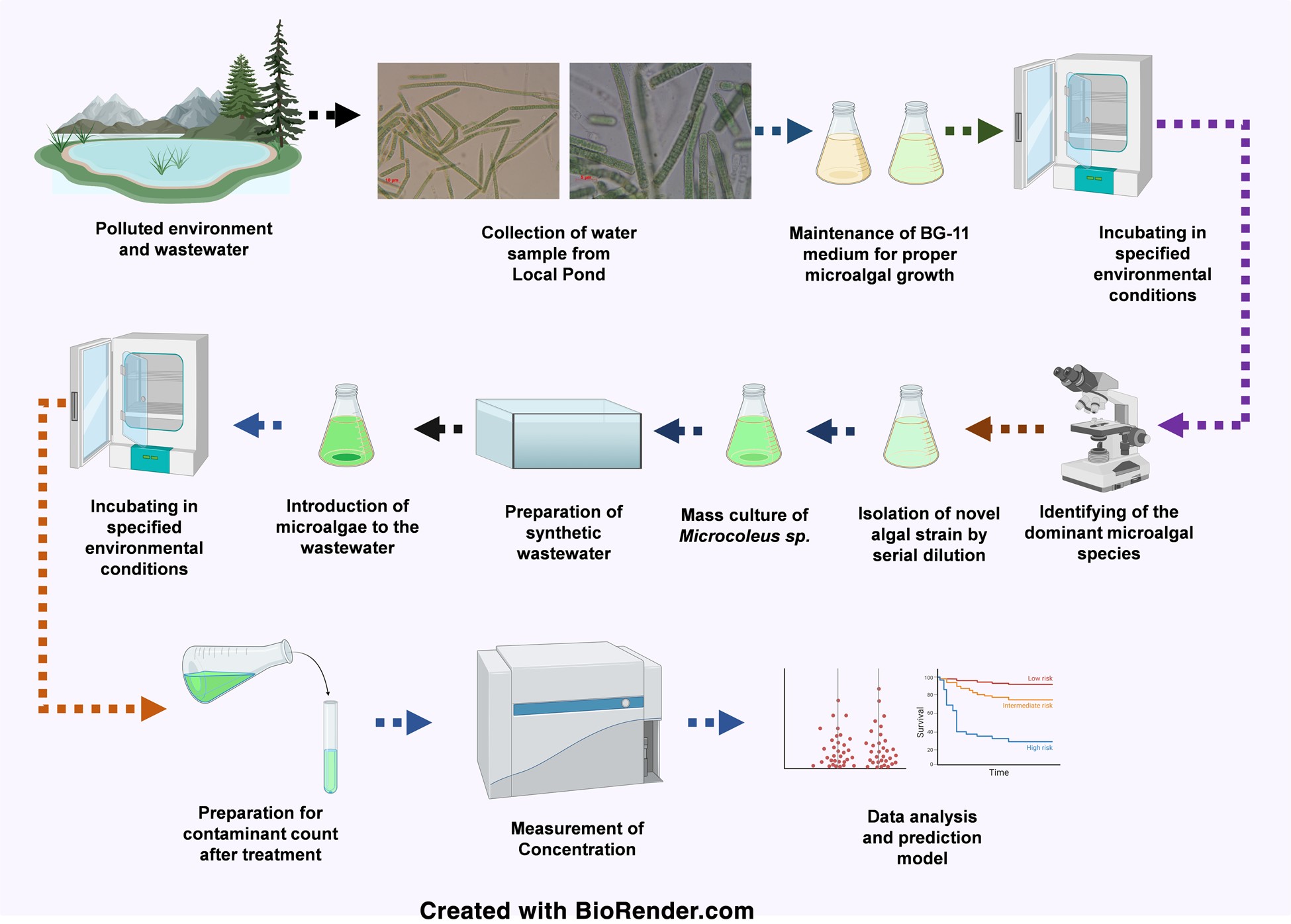
4. Microbial Bioremediation Using Cyanobacteria
Role: Research Assistant
Institute: Department of Civil & Environmental Engineering, SUST
Duration: November 2020 – March 2021
In this project, I cultivated blue-green algae (Microcoleus sp.) for use in biosorptive removal of heavy metals from industrial wastewater. I optimized culture conditions and conducted batch absorption assays under varying pH and retention times.
Analytical methods such as AAS and spectrophotometry were employed to calculate removal efficiencies. Data was modeled using Freundlich and Langmuir isotherms. This work contributed to:
- AQUA – Water Infrastructure, Ecosystems and Society (2023): Novel application of Microcoleus in bioremediation
