Since its emergence in late 2019, COVID-19, caused by the SARS-CoV-2 virus, has had a devastating global impact, with over 674 million confirmed cases and more than 6.7 million deaths reported by January 29, 2023. Like other RNA viruses, SARS-CoV-2 is prone to frequent mutations, and over time these changes have led to the appearance of new variants with distinct characteristics, including increased transmissibility, altered virulence, and the ability to evade immune protection from vaccination or prior infection, resulting in breakthrough cases and reinfections. To better understand this evolutionary process, we analyzed 10,531 whole-genome sequences of all major reported variants worldwide using computational tools such as NextCladeCLI, NextStrain, PROVEAN, PolyPhen-2, PredictSNP, and machine learning models to track, predict, and validate the impact of mutations. Compared to the original Wuhan-Hu-1 reference genome, we identified 16,954 mutations, with the Omicron variant showing the highest mutation load—6,307 mutations from 1,947 sequences, 67.8% of which were unique.
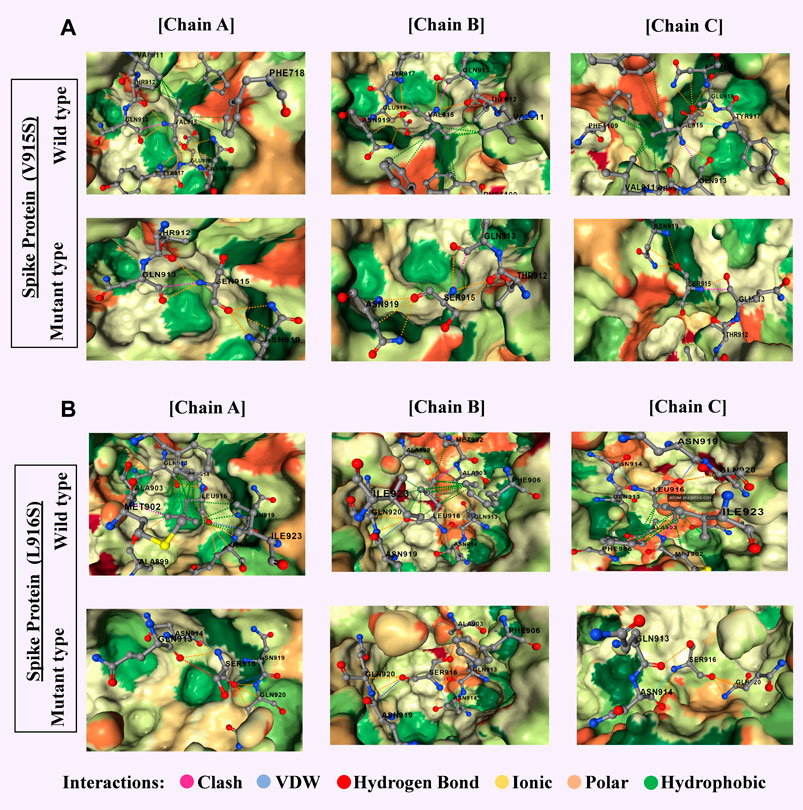
Omicron’s spike protein contained 876 mutations, including 443 predicted to be harmful, with 256 being unique non-synonymous changes; most were concentrated in the receptor-binding domain, which directly interacts with human cells, likely contributing to Omicron’s high transmissibility and ability to evade neutralizing antibodies. In contrast, the Delta variant, analyzed from 1,884 sequences, had 4,468 mutations, 66% unique, with many occurring in amino acid positions 911–924—regions associated with B-cell and T-cell immune recognition—indicating a different evolutionary focus on immune evasion rather than binding efficiency alone. These patterns suggest that while Omicron has evolved primarily to spread more efficiently, Delta’s changes targeted the immune system’s ability to detect and respond to it. The presence of persistent, unique, and harmful mutations in Omicron points to potential targets for updated vaccines or antiviral drug development, as focusing on these mutations could improve disease control and prevention strategies. Our findings emphasize the importance of continuous global genomic surveillance, which enables early detection of significant mutations and guides the timely adaptation of public health measures, vaccines, and treatments. SARS-CoV-2’s rapid and ongoing genetic evolution means that static solutions are insufficient; instead, a proactive approach combining real-time monitoring, advanced computational analysis, and targeted biomedical interventions is essential to staying ahead of the virus and mitigating its health, social, and economic impacts worldwide.

 Omicron and (B) Delta variants.jpg)