Research Projects
As a molecular biologist and bioinformatician working across both clinical and computational research, I have actively contributed to a range of interdisciplinary projects spanning infectious disease diagnostics, transcriptomics, RNA therapeutics, environmental virology, and AI-driven precision medicine. My project portfolio reflects a balance between wet-lab execution and advanced computational modeling—supported by collaborations in academia, government, and clinical trial networks.
Each project detailed below highlights not only the core scientific question but also my specific role in hypothesis development, data analysis, molecular assays, coding, and manuscript preparation. Several of these initiatives have led to peer-reviewed publications in high-impact journals and have directly informed my long-term research vision.
From uncovering mutational patterns in viral genomes to building deep learning models that predict regulatory RNA interactions, my work is centered on translating molecular data into real-world diagnostic and therapeutic solutions. This body of research also serves as a strong foundation for pursuing doctoral-level training in computational biology, systems medicine, and translational genomics.
1. Early Diagnosis and Risk Assessment of PROM in Rural Bangladesh
Role: Molecular Biologist
Institute: Advanced Molecular Lab, President Abdul Hamid Medical College & Hospital, Kishoreganj, Bangladesh
Duration: December 2023 – Present
Funding: Bangladesh Government Research Initiative
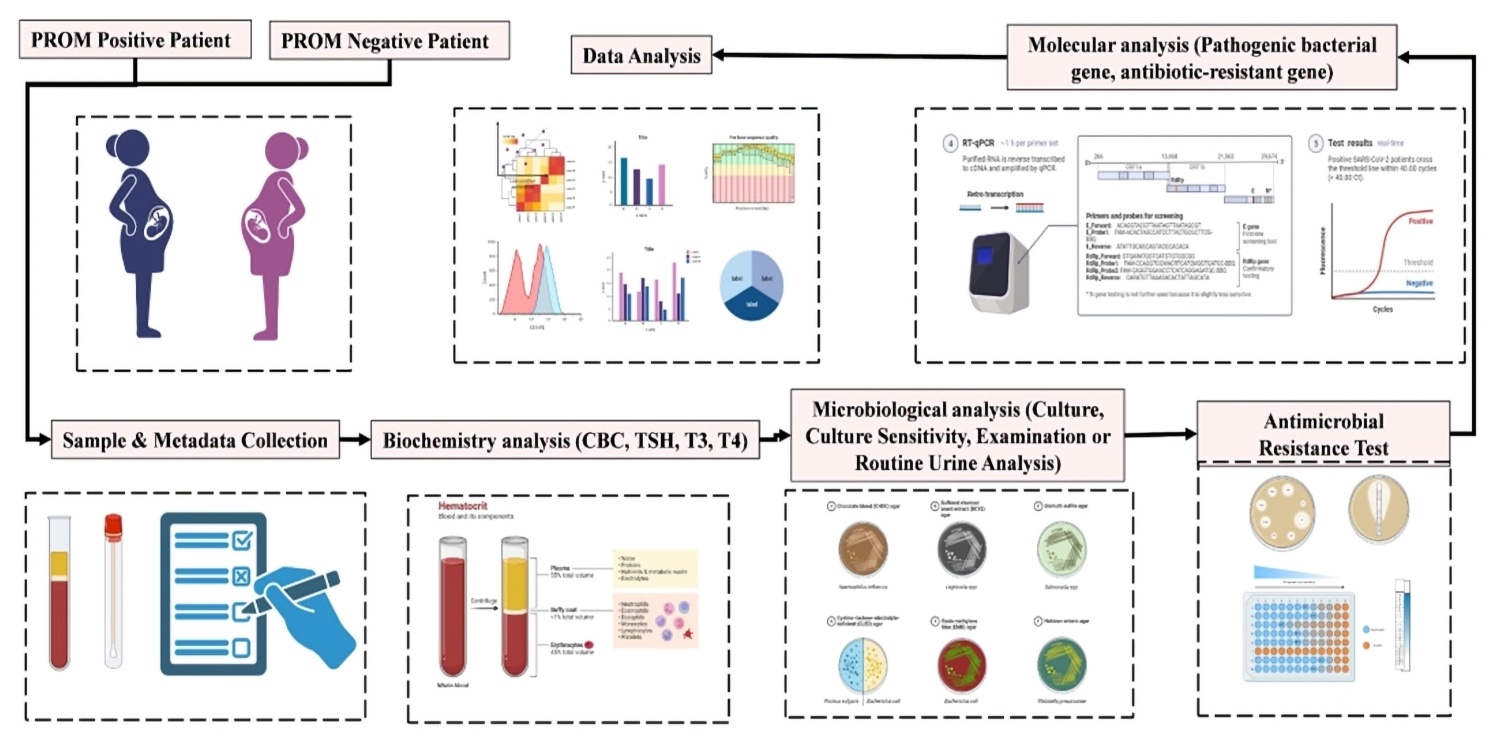
This government-funded multicenter study aims to develop early diagnostic models for identifying risk factors and molecular biomarkers associated with Premature Rupture of Membranes (PROM) in pregnant women, especially in resource-limited rural areas of Bangladesh. PROM is a significant cause of maternal and neonatal morbidity, and early detection is critical for prevention and intervention.
In this study, I oversee all molecular laboratory workflows, including patient sampling, RNA/DNA extraction, cDNA library preparation, qRT-PCR assay development, and ELISA-based protein detection. Additionally, I participate in the microbiological analysis of cervical and vaginal swabs to investigate potential infectious etiologies contributing to PROM cases.
One of the core innovations of this project is integrating clinical metadata with biomolecular signatures to establish a predictive diagnostic algorithm. My responsibilities also include developing data pipelines and visualization models in R and Python, applying statistical tests (t-tests, chi-square, ANOVA), and building risk stratification models using logistic regression and decision trees.
This project not only enhances maternal health research in Bangladesh but also builds bridges between fieldwork, wet-lab diagnostics, and computational data analysis for translational impact.
2. Clinical Data Management for Toxicological Phase III Trials

Role: Clinical Data Manager
Institute: Toxicology Society of Bangladesh, Dhaka Medical College Hospital, Bangladesh.
Duration: December 2023 – Present
I am currently engaged in managing clinical trial data for a Phase III government-monitored therapeutic intervention under the Toxicology Society of Bangladesh. This role involves overseeing every aspect of clinical data integrity, from electronic Case Report Form (eCRF) development to final statistical validation and reporting.
One of the primary tasks is the creation and optimization of a real-time interactive dashboard that tracks enrollment, intervention compliance, adverse events, and efficacy endpoints. This dashboard, built using Python Dash and Power BI, integrates directly with REDCap and SQL databases, enabling seamless data updates and quality control.
In addition to data infrastructure, I regularly audit and clean data, run multivariate and longitudinal analyses (using SPSS, R), and assist the clinical trial team in adapting protocols based on interim findings. I also coordinate the development of automated systems for real-time monitoring and validation of patient safety parameters, ensuring regulatory compliance and high trial fidelity.
This role has significantly strengthened my ability to work with large-scale clinical data systems and has deepened my understanding of statistical programming, biomedical informatics, and trial governance—essential skills for future PhD work in systems biology and translational data science.
3. Deep Learning Framework for Predicting miRNA–RRE Binding in HIV
Role: Lead Bioinformatician
Institute: Department of Biochemistry and Molecular Biology, Shahjalal University of Science and Technology (SUST), Sylhet, Bangladesh
Duration: February 2024 – Present
This project focuses on developing a deep learning–based predictive framework to identify and rank microRNA (miRNA) candidates that can effectively bind to the HIV Rev Response Element (RRE), particularly at the critical Stem IIB region. The project’s core objective is to discover potential miRNA-based therapeutic interventions that could disrupt Rev-mediated RNA nuclear export and replication of HIV.
The model integrates both sequence and structural features such as RNA secondary structure, minimum free energy (MFE), target site accessibility, GC content, and conservation scores. These features are extracted using RNAfold, ViennaRNA package, and RNAhybrid, while the deep learning pipeline is built using PyTorch and TensorFlow frameworks. Affinity scoring is further validated using experimentally backed datasets from microT-CDS and miRTarBase.
As part of the pipeline, multiple attention-based neural network models are being tested, including transformer architectures, to assess which configuration yields the most accurate miRNA–target site prediction. Each predicted miRNA candidate is ranked by predicted binding strength and biological feasibility, providing a list of vaccine-targetable synthetic miRNA analogs.
This project is particularly significant in the context of miRNA therapeutics, an emerging domain in antiviral treatment. It also aligns with global research directions in RNA therapeutics, artificial RNA design, and AI-assisted precision medicine.
4. Transcriptomic Analysis of Mouse Brain Cells at M11.5 for Neurodevelopmental Gene Expression

Role: Remote Research Assistant
Institute: Tulane University, Louisiana State, USA
Duration: April 2024 – Present
This project focuses on analyzing RNA-seq data from mouse brain tissue harvested at the embryonic day M11.5, a critical stage of neural development. The primary goal is to identify differential gene expression signatures associated with early neurogenesis, neuronal patterning, and transcriptional regulation during central nervous system formation.
Using the Galaxy server and command-line tools (FastQC, HISAT2, featureCounts, DESeq2), I preprocess raw sequencing data, align reads to the GRCm38 mouse genome, and quantify transcript abundance. This workflow is supported by statistical filtering, GO/KEGG enrichment analysis, and visualization using R (ggplot2, pheatmap, ClusterProfiler).
Preliminary findings have revealed key transcription factors and signaling pathways, such as Notch, Wnt, and Sonic Hedgehog, that exhibit stage-specific upregulation. These discoveries contribute to a deeper understanding of neurodevelopmental gene networks and provide insights into translational models of congenital brain disorders.
This project expands my multi-omics expertise and prepares me for integrative systems biology research at the doctoral level.
5. ML-Driven miRNA Therapeutics for Cancer Receptor Gene Suppression
Role: Principal Investigator (Solo Research)
Duration: January 2024 – Present
In this independent project, I am designing a deep learning model to identify optimized miRNA sequences that can silence overexpressed receptor genes associated with aggressive cancers, such as EGFR, HER2, and PDGFR. These receptors play pivotal roles in tumor proliferation, metastasis, and immune evasion.
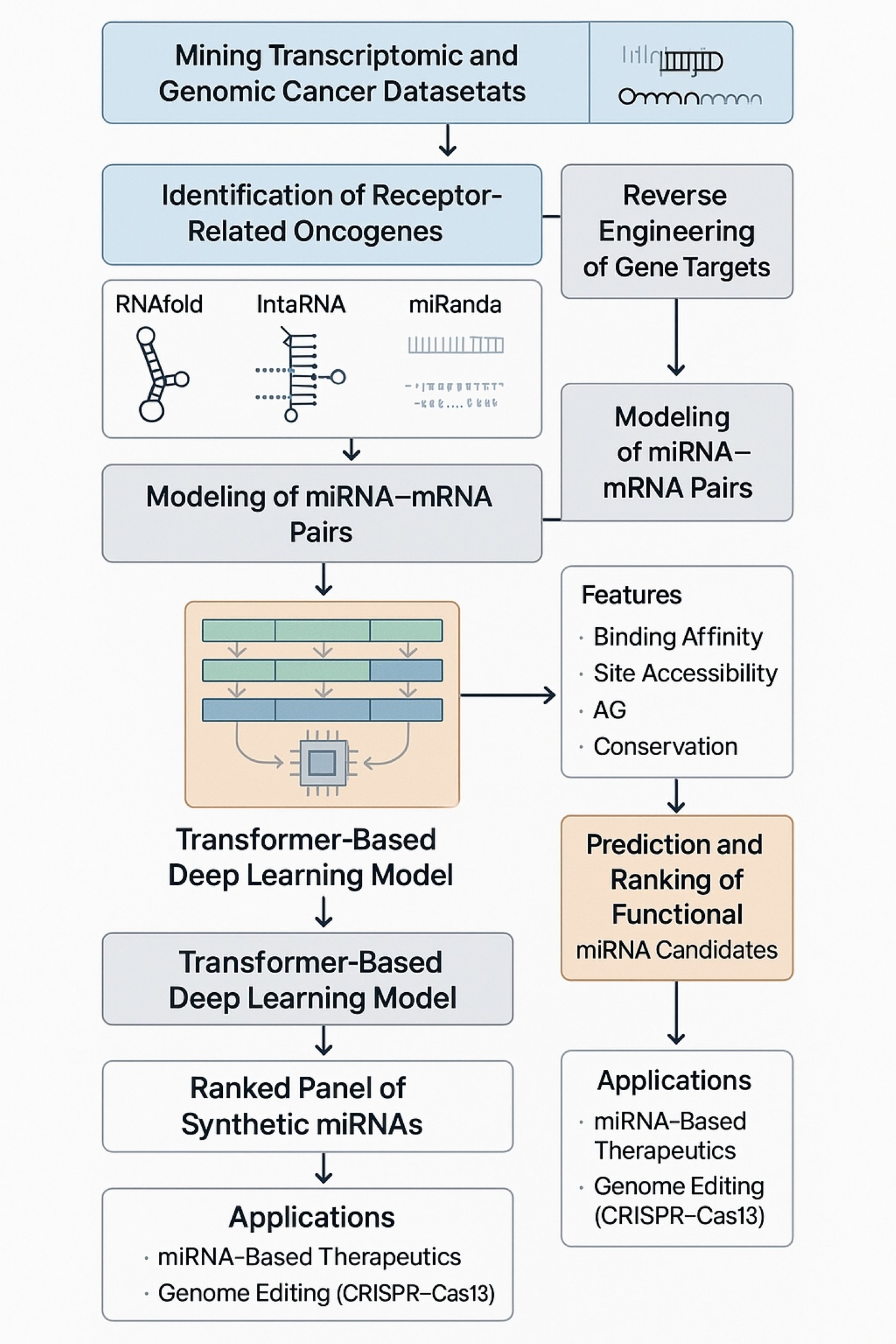
I begin by mining transcriptomic and genomic cancer datasets (TCGA, GTEx) to identify receptor-related oncogenes with aberrant expression. The selected gene targets are then reverse-engineered to trace their DNA transcriptional origins. I use RNAfold, IntaRNA, and miRanda to model secondary structures and base-pairing energetics of candidate miRNA–mRNA pairs.
On top of that, I’ve developed a custom transformer-based deep learning architecture that integrates features such as binding affinity, site accessibility, ∆G, and conservation scores to predict and rank functional miRNA candidates.
The final output will be a ranked panel of synthetic miRNAs with strong binding to key regulatory sequences upstream of these receptors, potentially applicable in miRNA-based therapeutics or genome editing (CRISPR–Cas13). This project represents the convergence of precision oncology, computational RNA biology, and AI—a nexus I aim to further explore in my PhD studies.

