Molecular Biologist
Molecular Diagnostics | Viral Genomics
Advanced Molecular Lab, PAHMCH
Zafrabad, Kishoreganj, Bangladesh
Bioinformatician
Deep Learning in Translational Biology
Department of Biochemistry and Molecular Biology, Shahjalal University of Science and Technology
Sylhet-3114, Bangladesh
Clinical Data Manager
Toxicology Society of Bangladesh
Dhaka Medical College Hospital
Dhaka-1000, Bangladesh
Greetings! This is Marzan, a Molecular Biologist and Bioinformatician specializing in infectious disease diagnostics, viral genomics, and ML-driven translational biology. My work bridges molecular-level experimentation and computational analysis to develop predictive and actionable diagnostic strategies.
Currently, I serve in three parallel research positions: as a Clinical Data Manager at Dhaka Medical College Hospital, a Molecular Researcher at President Abdul Hamid Medical College and Hospital, and a Bioinformatics Researcher at Shahjalal University of Science and Technology. Through these complementary roles, I contribute to next-generation diagnostic innovation, structural modeling, and the integration of multi-omic data.
My research focuses on the intersection of RNA biology, molecular diagnostics, and machine learning in genomic medicine. I am particularly interested in modeling miRNA–mRNA interactions by integrating RNA secondary structure, thermodynamic features, and sequence-based targeting rules to uncover regulatory mechanisms involved in cancer, immune modulation, and therapeutic response.
In parallel, I explore multi-omics integration and molecular dynamics simulation to discover functional biomarkers and design intelligent diagnostic systems. By combining deep learning with biological interpretability, my goal is to create scalable, AI-powered platforms for early disease detection and precision healthcare.
As a molecular biologist and bioinformatician working across both clinical and computational research, I have actively contributed to a range of interdisciplinary projects spanning infectious disease diagnostics, transcriptomics, RNA therapeutics, environmental virology, and AI-driven precision medicine. My project portfolio reflects a balance between wet-lab execution and advanced computational modeling—supported by collaborations in academia, government, and clinical trial networks.
Each project detailed below highlights not only the core scientific question but also my specific role in hypothesis development, data analysis, molecular assays, coding, and manuscript preparation. Several of these initiatives have led to peer-reviewed publications in high-impact journals and have directly informed my long-term research vision.
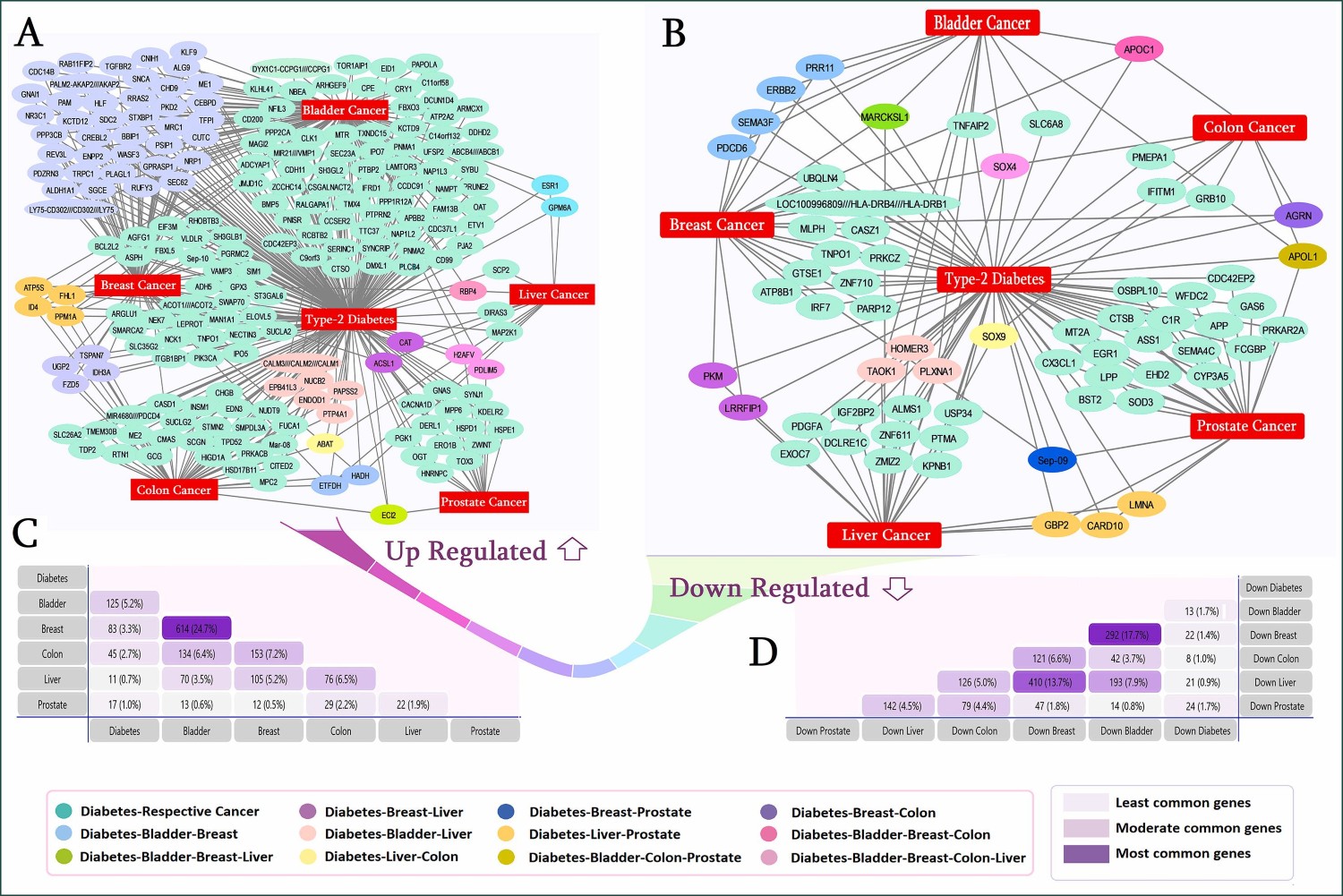
- A. A. Marzan et al., ‘Probing biological network in concurrent carcinomas and Type-2 diabetes for potential biomarker screening: An advanced computational paradigm’, Advances in Biomarker Sciences and Technology, vol. 5, Oct. 2023.
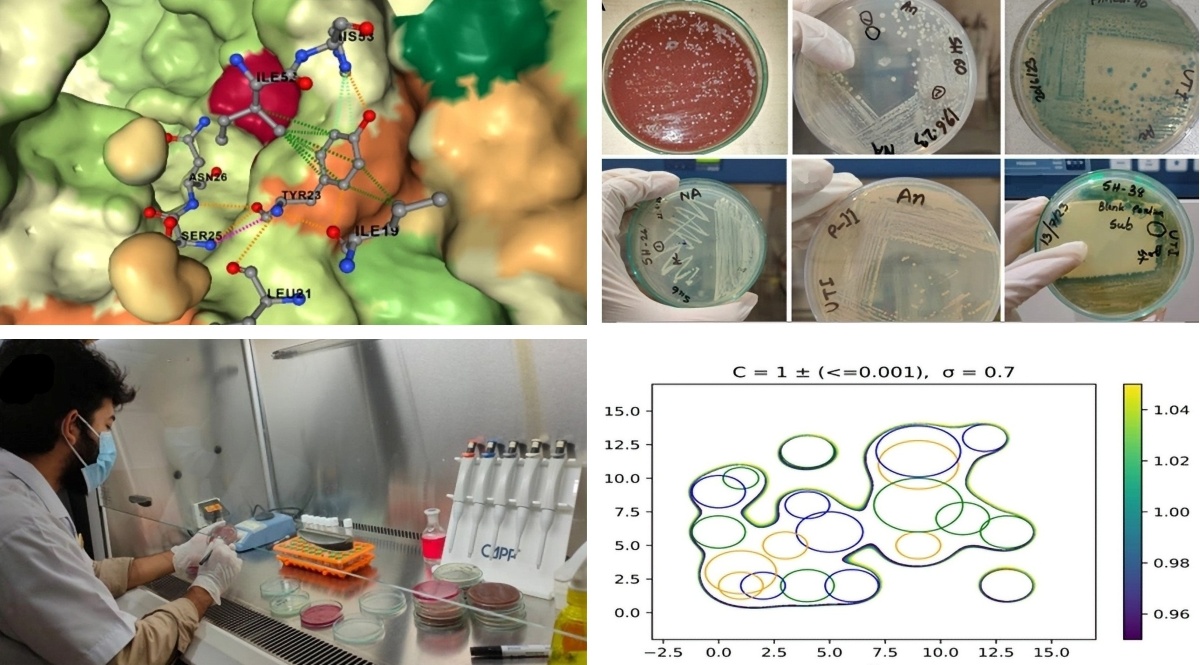
- Md. A. Islam et al., ‘Variant-specific deleterious mutations in the SARS-CoV-2 genome reveal immune responses and potentials for prophylactic vaccine development’, Front Pharmacol, vol. 14, Feb. 2023.
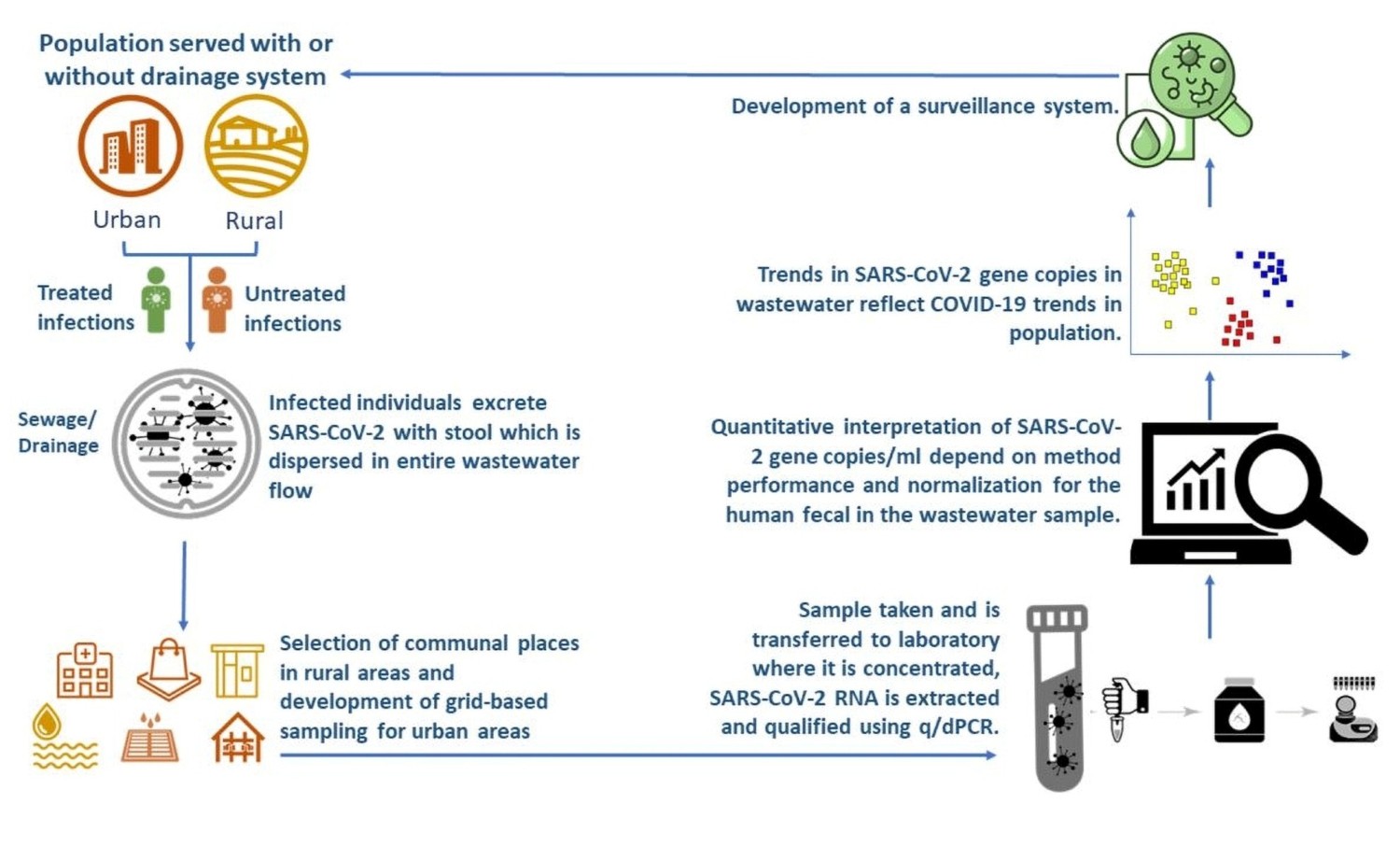
- M. Jakariya et al., ‘Wastewater-based epidemiological surveillance to monitor the prevalence of SARS-CoV-2 in developing countries with onsite sanitation facilities’, Environmental Pollution, vol. 311, Oct. 2022.
My expertise bridges wet-lab experimentation with computational precision, enabling me to extract biological meaning from complex datasets and translate them into actionable research insights.
From viral genome analysis and biomarker discovery to deep learning applications for RNA sequence targeting, I’ve developed robust skills in both molecular biology techniques (qRT-PCR, ELISA, biomarker profiling) and computational pipelines (RNAfold, AutoDock, RNA-seq, GROMACS).
I hold professional certifications in Python, R programming, and advanced data science tools, allowing me to build predictive models for miRNA–mRNA interactions, mutation impact profiling, and transcriptomic diagnostics.



.jpg)